MATLAB Tutorial
Welcome to the MATLAB tutorial version of Data Science Rosetta Stone. Before beginning this tutorial, please check to make sure you have MATLAB installed.
Note: In MATLAB,
% This is a single line comment. %{ This is a paragraph comment %}
Now let’s get started!
1 Reading in Data and Basic Statistical Functions
1.1 Read in the data.
a) Read the data in as a .csv file.
First specify the format of the variables to be read in: %C = character, %d = integer, %f = floating point number.
formatSpec = '%C%C%d%f%f'; student = readtable('class.csv', 'Delimiter', ',', 'Format', formatSpec);
b) Read the data in as a .xls file.
MATLAB reads tables from .xlsx formats, which is another version of an Excel file.
student_xlsx = readtable('class.xlsx');
c) Read the data in as a .json file.
student_json = jsondecode(fileread('class.json'));
1.2 Find the dimensions of the data set.
1.3 Find basic information about the data set.
summary(student);
Variables:
Name: 19×1 categorical
Values:
Alfred 1
Alice 1
Barbara 1
Carol 1
Henry 1
James 1
Jane 1
Janet 1
Jeffrey 1
John 1
Joyce 1
Judy 1
Louise 1
Mary 1
Philip 1
Robert 1
Ronald 1
Thomas 1
William 1
Sex: 19×1 categorical
Values:
F 9
M 10
Age: 19×1 int32
Values:
Min 11
Median 13
Max 16
Height: 19×1 double
Values:
Min 51.3
Median 62.8
Max 72
Weight: 19×1 double
Values:
Min 50.5
Median 99.5
Max 150
summary()
1.4 Look at the first 5 (last 5) observations.
The ":" operator tells MATLAB to print all columns (variables), while "1:5" indicates to print only the first 5 observations.
disp(student(1:5,:));
Name Sex Age Height Weight
_______ ___ ___ ______ ______
Alfred M 14 69 112.5
Alice F 13 56.5 84
Barbara F 13 65.3 98
Carol F 14 62.8 102.5
Henry M 14 63.5 102.5
disp()
1.5 Calculate means of numeric variables.
age = table2array(student(:,3)); disp(mean(age));
13.3158
height = table2array(student(:,4)); disp(mean(height));
62.3368
weight = table2array(student(:,5)); disp(mean(weight));
100.0263table2array() | mean() | disp()
1.6 Compute summary statistics of the data set.
numeric_vars = student(:,{'Age', 'Height', 'Weight'});
statarray = grpstats(numeric_vars, [], {'min', 'median', 'mean', 'max'});
disp(statarray);
GroupCount min_Age median_Age mean_Age max_Age min_Height median_Height mean_Height max_Height min_Weight median_Weight mean_Weight max_Weight
__________ _______ __________ ________ _______ __________ _____________ ___________ __________ __________ _____________ ___________ __________
All 19 11 13 13.316 16 51.3 62.8 62.337 72 50.5 99.5 100.03 150
grpstats() | disp()
1.7 Descriptive statistics functions applied to variables of the data set.
weight = table2array(student(:,5)); disp(std(weight));
22.7739
disp(sum(weight));
1.9005e+03
disp(length(weight));
19
disp(max(weight));
150
disp(min(weight));
50.5000
disp(median(weight));
99.5000table2array() | std() | sum() | length() | max() | min() | median()
1.8 Produce a one-way table to describe the frequency of a variable.
a) Produce a one-way table of a discrete variable.
tabulate(age);
Value Count Percent
1 0 0.00%
2 0 0.00%
3 0 0.00%
4 0 0.00%
5 0 0.00%
6 0 0.00%
7 0 0.00%
8 0 0.00%
9 0 0.00%
10 0 0.00%
11 2 10.53%
12 5 26.32%
13 3 15.79%
14 4 21.05%
15 4 21.05%
16 1 5.26%
tabulate()
b) Produce a one-way table of a categorical variable.
sex = table2array(student(:,{'Sex'}));
tabulate(sex);
Value Count Percent
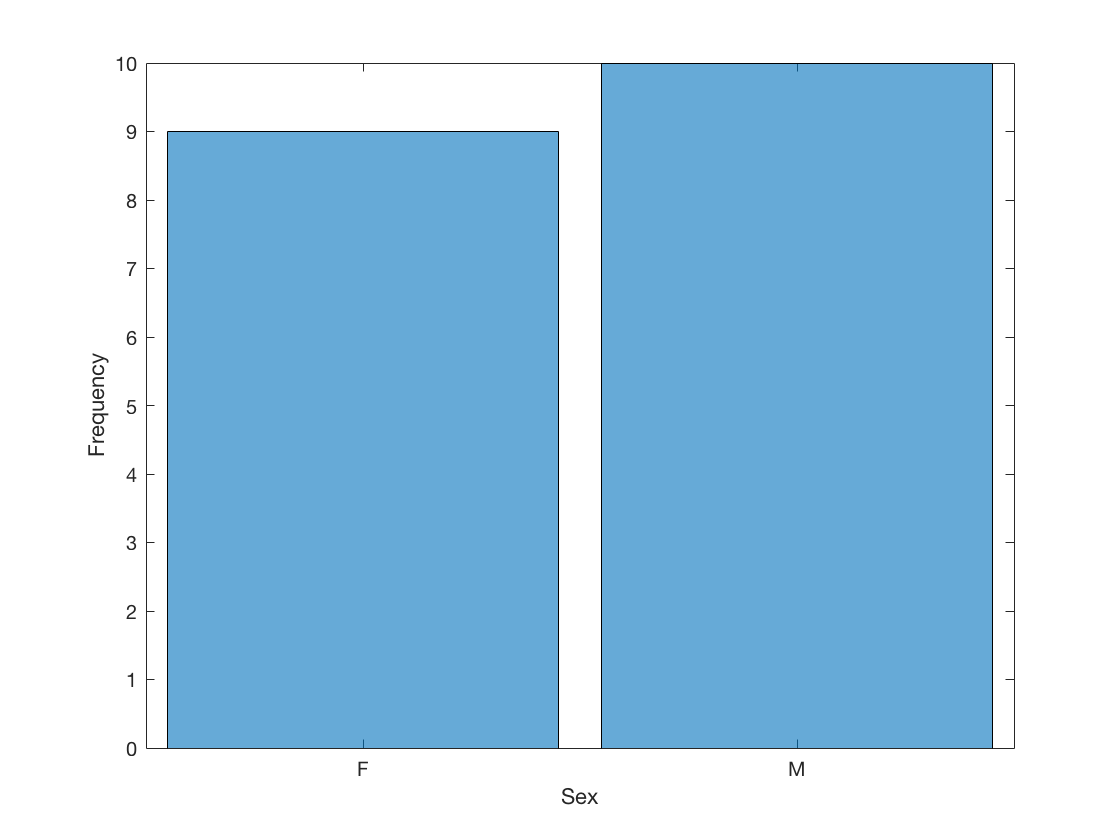
F 9 47.37%
M 10 52.63%
1.9 Produce a two-way table to describe the frequency of two categorical or discrete variables.
crosstable = varfun(@(x) length(x), student, 'GroupingVariables', {'Age' 'Sex'}, 'InputVariables', {}); disp(crosstable);
Age Sex GroupCount
___ ___ __________
11 F 1
11 M 1
12 F 2
12 M 3
13 F 2
13 M 1
14 F 2
14 M 2
15 F 2
15 M 2
16 M 1
varfun()
1.10 Select a subset of the data that meets a certain criterion.
% Find the indices of those students who are females, and then get those observations % from the student data frame. females = student(student.Sex == 'F',:); disp(females(1:5,:));
Name Sex Age Height Weight
_______ ___ ___ ______ ______
Alice F 13 56.5 84
Barbara F 13 65.3 98
Carol F 14 62.8 102.5
Jane F 12 59.8 84.5
Janet F 15 62.5 112.5
1.11 Determine the correlation between two continuous variables.
The first argument of the cat function is dim, which is specified as 2 here to indicate to concatenate column-wise.
height_weight = cat(2,table2array(student(:,4)),table2array(student(:,5))); disp(corr(height_weight));
1.0000 0.8778
0.8778 1.0000
table2array() | cat() | corr()
2 Basic Graphing and Plotting Functions
2.1 Visualize a single continuous variable by producing a histogram.
Weight = table2array(student(:,{'Weight'}));
histogram(Weight, 40:20:160)
xlabel('Weight');
ylabel('Frequency');
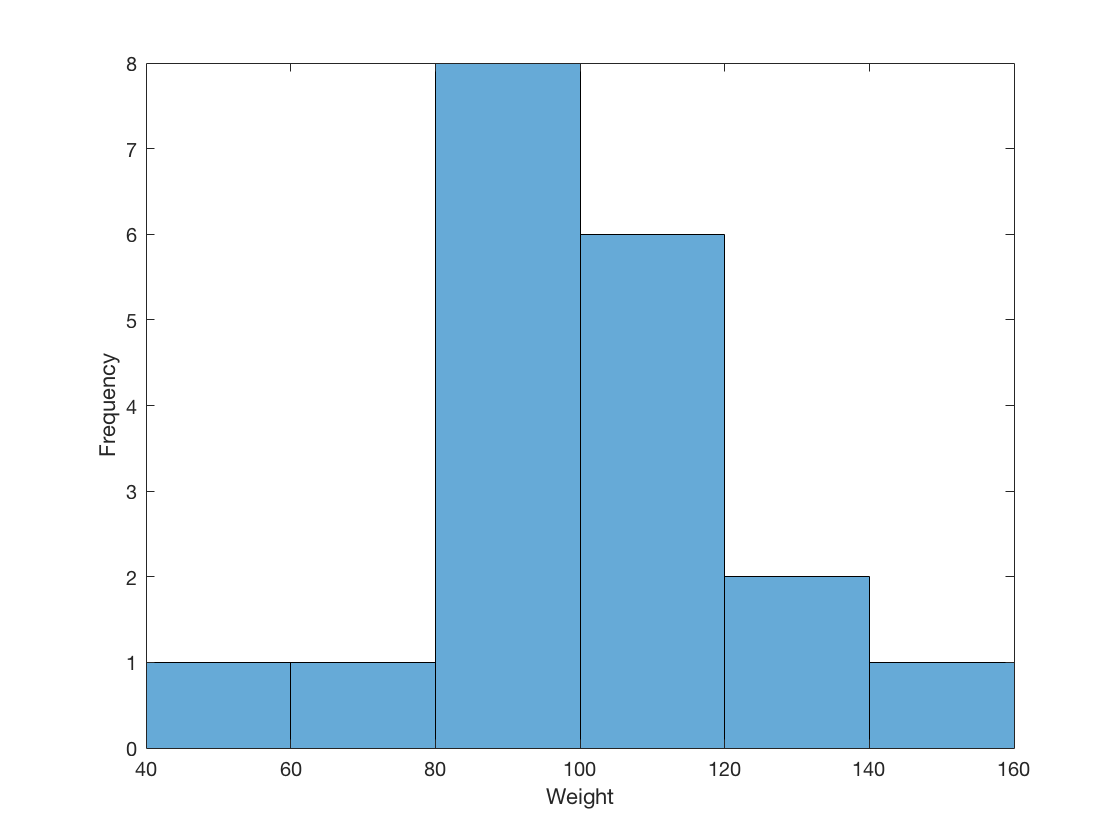
2.2 Visualize a single continuous variable by producing a boxplot.
boxplot(Weight); mx = mean(Weight); ylabel('Weight'); hold on plot(mx, 'd') hold off
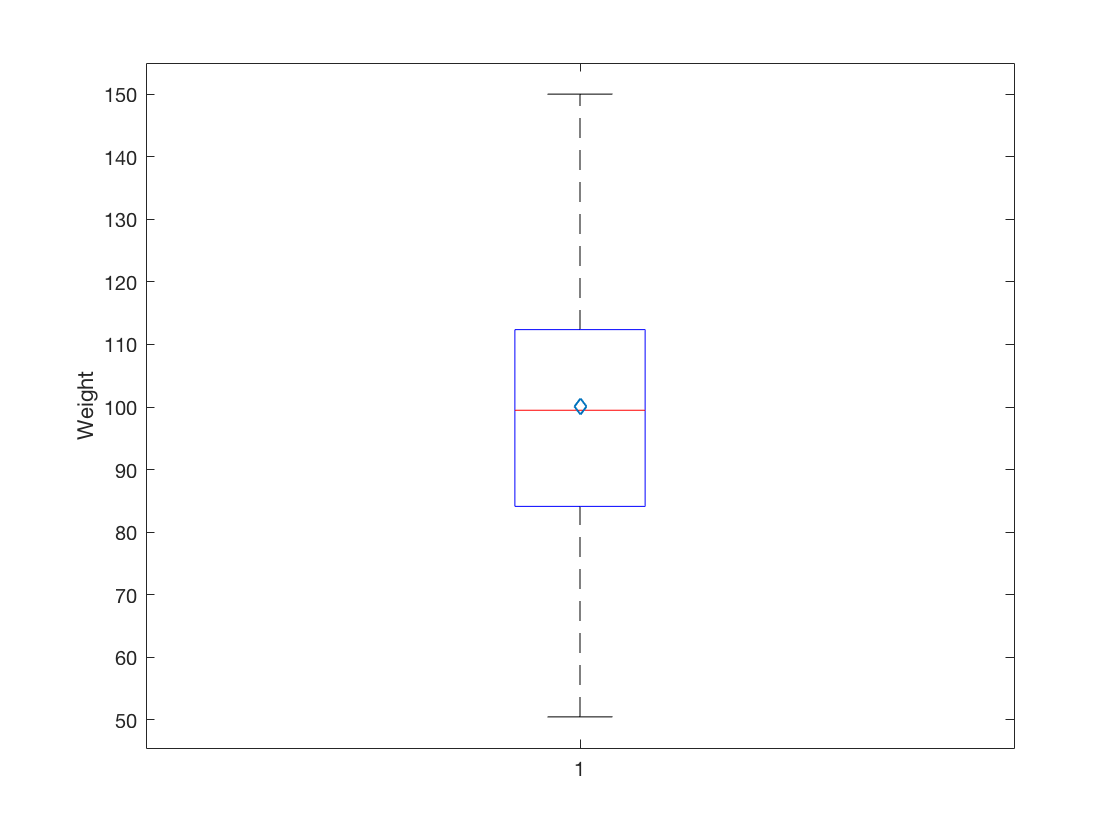
2.3 Visualize two continuous variables by producing a scatterplot.
Height = table2array(student(:,{'Height'}));
scatter(Height, Weight)
xticks(50:5:75)
yticks(40:20:160)
xlabel('Height')
ylabel('Weight')
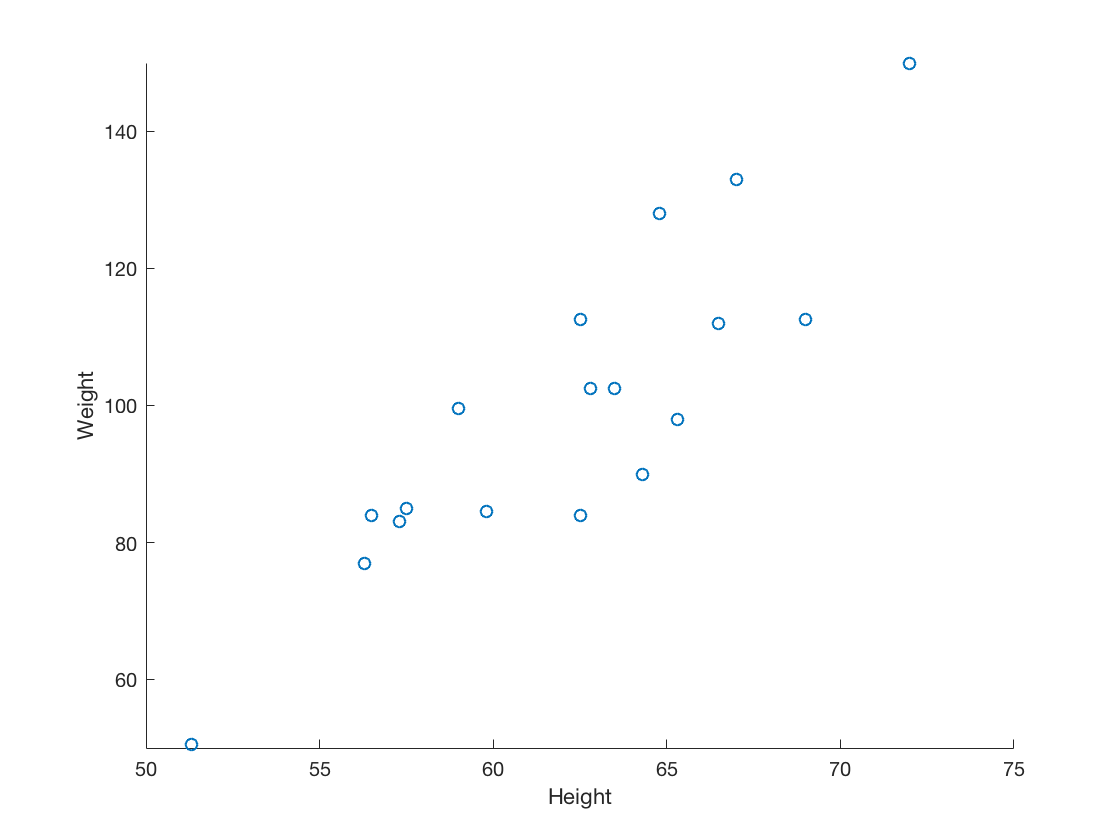
2.4 Visualize a relationship between two continuous variables by producing a scatterplot and a plotted line of best fit.
scatter(Height, Weight) xticks(50:5:75) yticks(40:20:160) xlabel('Height') ylabel('Weight') b = polyfit(Height, Weight,1); m = b(1); y_int = b(2); lsline annotation('textbox', [.2 .5 .3 .3], 'String', sprintf('Line: y = %fx + %f', m, y_int), ... 'FitBoxToText', 'on');
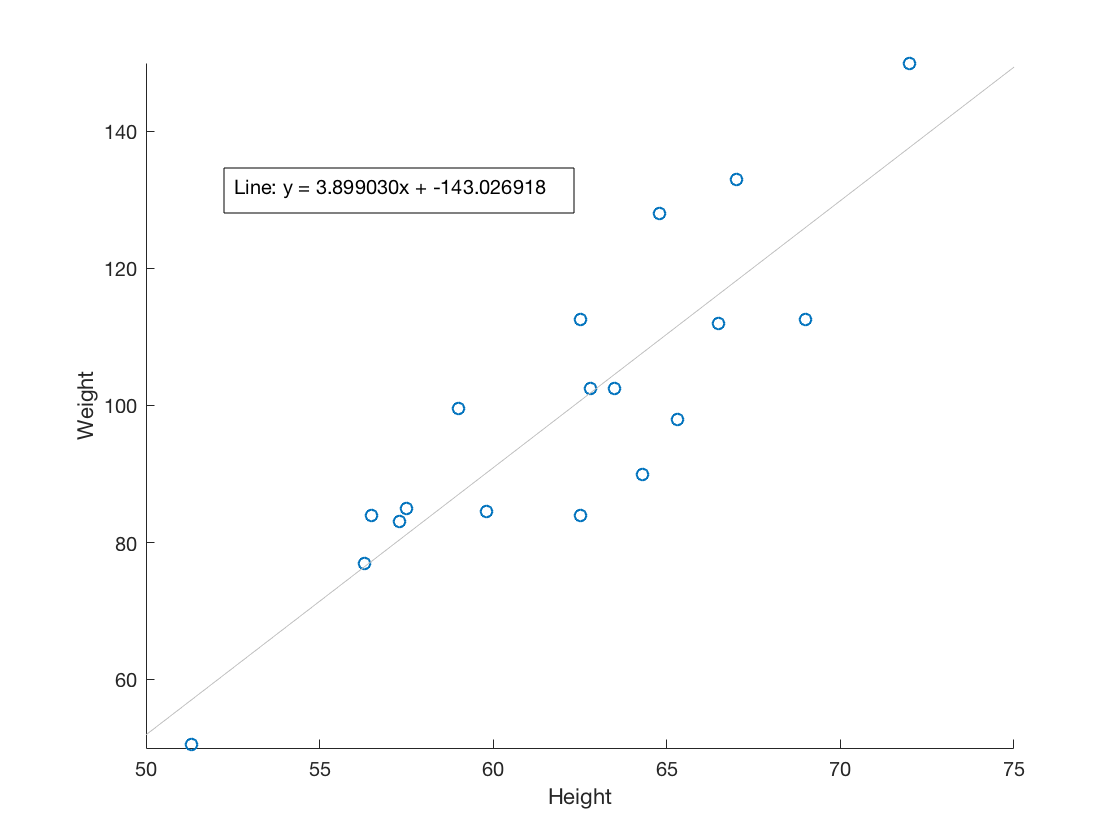
2.5 Visualize a categorical variable by producing a bar chart.
Sex = table2array(student(:,{'Sex'}));
histogram(Sex)
xlabel('Sex')
ylabel('Frequency')
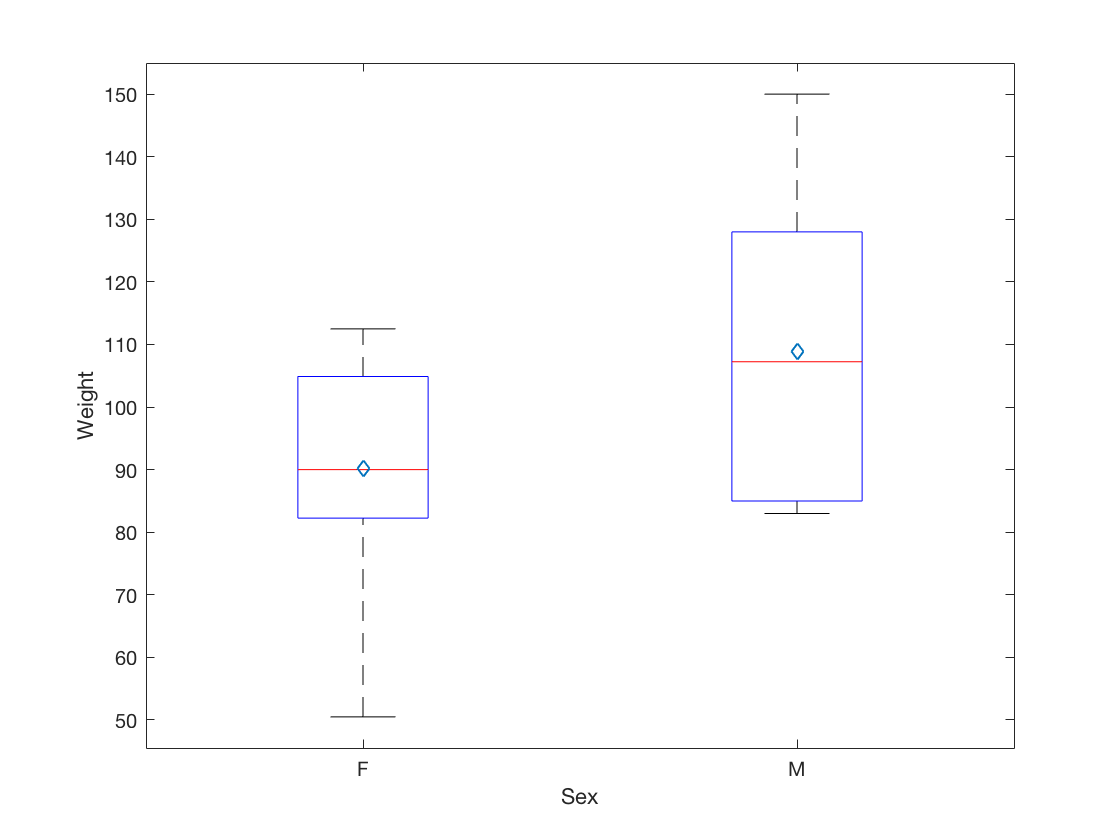
2.6 Visualize a continuous variable, grouped by a categorical variable, by producing side-by-side boxplots.
females = student(student.Sex == 'F',:); males = student(student.Sex == 'M',:); Female_Weight = table2array(females(:,{'Weight'})); Male_Weight = table2array(males(:,{'Weight'})); clf boxplot(Weight, Sex); means = [mean(Female_Weight), mean(Male_Weight)]; xlabel('Sex'); ylabel('Weight'); hold on plot(means, 'd') hold off
3 Basic Data Wrangling and Manipulation
3.1 Create a new variable in a data set as a function of existing variables in the data set.
% The "./" (and similarly, ".^2") tells MATLAB to divide (and similarly, exponentiate) % element-wise, instead of matrix-wise. student.BMI = student.Weight ./ student.Height .^ 2 * 703; disp(student(1:5,:));
Name Sex Age Height Weight BMI
_______ ___ ___ ______ ______ ______
Alfred M 14 69 112.5 16.612
Alice F 13 56.5 84 18.499
Barbara F 13 65.3 98 16.157
Carol F 14 62.8 102.5 18.271
Henry M 14 63.5 102.5 17.87
3.2 Create a new variable in a data set using if/else logic of existing variables in the data set.
student.BMI_Class = student.Name; for i = 1:size(student,1) if student.BMI(i) < 19.0 student.BMI_Class(i) = 'Underweight'; else student.BMI_Class(i) = 'Healthy'; end end disp(student(1:5,:));
Name Sex Age Height Weight BMI BMI_Class
_______ ___ ___ ______ ______ ______ ___________
Alfred M 14 69 112.5 16.612 Underweight
Alice F 13 56.5 84 18.499 Underweight
Barbara F 13 65.3 98 16.157 Underweight
Carol F 14 62.8 102.5 18.271 Underweight
Henry M 14 63.5 102.5 17.87 Underweight
3.3 Create new variables in a data set using mathematical functions applied to existing variables in the data set.
student.LogWeight = log(student.Weight); student.ExpAge = exp(double(student.Age)); student.SqrtHeight = sqrt(student.Height); student.BMI_Neg = student.BMI; for i = 1:size(student,1) if student.BMI(i) < 19.0 student.BMI_Neg(i) = -student.BMI(i); end end student.BMI_Pos = abs(student.BMI_Neg); student.BMI_Check = (student.BMI_Pos == student.BMI); disp(student(1:5,:));
Name Sex Age Height Weight BMI BMI_Class LogWeight ExpAge SqrtHeight BMI_Neg BMI_Pos BMI_Check
_______ ___ ___ ______ ______ ______ ___________ _________ __________ __________ _______ _______ _________
Alfred M 14 69 112.5 16.612 Underweight 4.723 1.2026e+06 8.3066 -16.612 16.612 true
Alice F 13 56.5 84 18.499 Underweight 4.4308 4.4241e+05 7.5166 -18.499 18.499 true
Barbara F 13 65.3 98 16.157 Underweight 4.585 4.4241e+05 8.0808 -16.157 16.157 true
Carol F 14 62.8 102.5 18.271 Underweight 4.6299 1.2026e+06 7.9246 -18.271 18.271 true
Henry M 14 63.5 102.5 17.87 Underweight 4.6299 1.2026e+06 7.9687 -17.87 17.87 true
log() | exp() | sqrt() | abs()
3.4 Drop variables from a data set.
Setting the variables to an empty array deletes the variables from the dataset.
student.LogWeight = []; student.ExpAge = []; student.SqrtHeight = []; student.BMI_Neg = []; student.BMI_Pos = []; student.BMI_Check = []; disp(student(1:5,:));
Name Sex Age Height Weight BMI BMI_Class
_______ ___ ___ ______ ______ ______ ___________
Alfred M 14 69 112.5 16.612 Underweight
Alice F 13 56.5 84 18.499 Underweight
Barbara F 13 65.3 98 16.157 Underweight
Carol F 14 62.8 102.5 18.271 Underweight
Henry M 14 63.5 102.5 17.87 Underweight
3.5 Sort a data set by a variable.
a) Sort data set by a continuous variable.
student = sortrows(student, 'Age');
disp(student(1:5,:));
Name Sex Age Height Weight BMI BMI_Class
______ ___ ___ ______ ______ ______ ___________
Joyce F 11 51.3 50.5 13.49 Underweight
Thomas M 11 57.5 85 18.073 Underweight
James M 12 57.3 83 17.772 Underweight
Jane F 12 59.8 84.5 16.612 Underweight
John M 12 59 99.5 20.094 Healthy
sortrows()
b) Sort data set by a categorical variable.
student = sortrows(student, 'Sex');
disp(student(1:5,:));
Name Sex Age Height Weight BMI BMI_Class
_______ ___ ___ ______ ______ ______ ___________
Joyce F 11 51.3 50.5 13.49 Underweight
Jane F 12 59.8 84.5 16.612 Underweight
Louise F 12 56.3 77 17.078 Underweight
Alice F 13 56.5 84 18.499 Underweight
Barbara F 13 65.3 98 16.157 Underweight
sortrows()
3.6 Compute descriptive statistics of continuous variables, grouped by a categorical variable.
group_means = grpstats(student, 'Sex', 'mean', 'DataVars', {'Age', 'Height', 'Weight', 'BMI'}); disp(group_means);
Sex GroupCount mean_Age mean_Height mean_Weight mean_BMI
___ __________ ________ ___________ ___________ ________
F F 9 13.222 60.589 90.111 17.051
M M 10 13.4 63.91 108.95 18.594
grpstats()
3.7 Add a new row to the bottom of a data set.
disp(student(15:19,:));
Name Sex Age Height Weight BMI BMI_Class
_______ ___ ___ ______ ______ ______ ___________
Alfred M 14 69 112.5 16.612 Underweight
Henry M 14 63.5 102.5 17.87 Underweight
Ronald M 15 67 133 20.828 Healthy
William M 15 66.5 112 17.805 Underweight
Philip M 16 72 150 20.341 Healthy
newObs = {'Name', 'Sex', 'Age', 'Height', 'Weight', 'BMI', 'BMI_Class';
'Jane', 'F', 14, 56.3, 77.0, 17.077695, 'Underweight'};
newTable = dataset2table(cell2dataset(newObs));
student = vertcat(student,newTable);
disp(student(16:20,:));
Name Sex Age Height Weight BMI BMI_Class
_______ ___ ___ ______ ______ ______ ___________
Henry M 14 63.5 102.5 17.87 Underweight
Ronald M 15 67 133 20.828 Healthy
William M 15 66.5 112 17.805 Underweight
Philip M 16 72 150 20.341 Healthy
Jane F 14 56.3 77 17.078 Underweight
cell2dataset() | dataset2table() | vertcat()
3.8 Create a user-defined function and apply it to a variable in the data set to create a new variable in the data set.
% To create a user-defined function, create a new file in MATLAB with the function definition, % and save the file as the function_name.m. Here, toKG.m would be: % % function KG = toKG(lb); % KG = 0.45359237 * lb; % end student.Weight_KG = toKG(student.Weight); disp(student(1:5,:));
Name Sex Age Height Weight BMI BMI_Class Weight_KG
_______ ___ ___ ______ ______ ______ ___________ _________
Joyce F 11 51.3 50.5 13.49 Underweight 22.906
Jane F 12 59.8 84.5 16.612 Underweight 38.329
Louise F 12 56.3 77 17.078 Underweight 34.927
Alice F 13 56.5 84 18.499 Underweight 38.102
Barbara F 13 65.3 98 16.157 Underweight 44.452
user-defined function
4 More Advanced Data Wrangling
4.1 Drop observations with missing information.
formatSpec = '%C%f%f%f%f%f%f'; fish = readtable('fish.csv', 'Delimiter', ',', ... 'Format', formatSpec); fish = sortrows(fish, 'Weight', 'descend'); disp(fish(1:5,:));
Species Weight Length1 Length2 Length3 Height Width
_______ ______ _______ _______ _______ ______ ______
Bream NaN 29.5 32 37.3 13.913 5.0728
Pike 1650 59 63.4 68 10.812 7.48
Pike 1600 56 60 64 9.6 6.144
Pike 1550 56 60 64 9.6 6.144
Pike 1250 52 56 59.7 10.686 6.9849
fish = rmmissing(fish); disp(fish(1:5,:));
Species Weight Length1 Length2 Length3 Height Width
_______ ______ _______ _______ _______ ______ ______
Pike 1650 59 63.4 68 10.812 7.48
Pike 1600 56 60 64 9.6 6.144
Pike 1550 56 60 64 9.6 6.144
Pike 1250 52 56 59.7 10.686 6.9849
Perch 1100 39 42 44.6 12.8 6.8684
readtable() | sortrows() | rmmissing()
4.2 Merge two data sets together on a common variable.
a) First, select specific columns of a data set to create two smaller data sets.
formatSpec = '%C%C%d%f%f'; student = readtable('class.csv', 'Delimiter', ',', ... 'Format', formatSpec); student1 = student(:, {'Name', 'Sex', 'Age'}); disp(student1(1:5,:));
Name Sex Age
_______ ___ ___
Alfred M 14
Alice F 13
Barbara F 13
Carol F 14
Henry M 14
student2 = student(:, {'Name', 'Height', 'Weight'});
disp(student2(1:5,:));
Name Height Weight
_______ ______ ______
Alfred 69 112.5
Alice 56.5 84
Barbara 65.3 98
Carol 62.8 102.5
Henry 63.5 102.5
readtable()
b) Second, we want to merge the two smaller data sets on the common variable.
new = join(student1, student2); disp(new(1:5,:));
Name Sex Age Height Weight
_______ ___ ___ ______ ______
Alfred M 14 69 112.5
Alice F 13 56.5 84
Barbara F 13 65.3 98
Carol F 14 62.8 102.5
Henry M 14 63.5 102.5
join()
c) Finally, we want to check to see if the merged data set is the same as the original data set.
4.3 Merge two data sets together by index number only.
a) First, select specific columns of a data set to create two smaller data sets.
newstudent1 = student(:, {'Name', 'Sex', 'Age'});
disp(newstudent1(1:5,:));
Name Sex Age
_______ ___ ___
Alfred M 14
Alice F 13
Barbara F 13
Carol F 14
Henry M 14
newstudent2 = student(:, {'Height', 'Weight'});
disp(newstudent2(1:5,:));
Height Weight
______ ______
69 112.5
56.5 84
65.3 98
62.8 102.5
63.5 102.5
b) Second, we want to join the two smaller data sets.
new2 = [newstudent1, newstudent2]; disp(new2(1:5,:));
Name Sex Age Height Weight
_______ ___ ___ ______ ______
Alfred M 14 69 112.5
Alice F 13 56.5 84
Barbara F 13 65.3 98
Carol F 14 62.8 102.5
Henry M 14 63.5 102.5
c) Finally, we want to check to see if the joined data set is the same as the original data set.
4.4 Create a pivot table to summarize information about a data set.
% Currently there is not a MATLAB function for creating pivot tables, but only user-defined functions % that could be used to create pivot tables.pivottable user-defined function
4.5 Return all unique values from a text variable.
price = readtable('price.xlsx');
disp(unique(price.STATE));
'Baja California Norte'
'British Columbia'
'California'
'Campeche'
'Colorado'
'Florida'
'Illinois'
'Michoacan'
'New York'
'North Carolina'
'Nuevo Leon'
'Ontario'
'Quebec'
'Saskatchewan'
'Texas'
'Washington'
readtable() | unique()5 Preparation & Basic Regression
5.1 Pre-process a data set using principal component analysis.
formatSpec = '%f%f%f%f%d'; iris = readtable('iris.csv', 'Delimiter', ',', ... 'Format', formatSpec); features = table2array(iris(:, {'SepalLength', 'SepalWidth', 'PetalLength', 'PetalWidth'})); % Z-score function to scale Zsc = @(x) (x-mean(x))./std(x); features_scaled = Zsc(features); disp(pca(features_scaled));
0.5224 0.3723 0.7210 -0.2620
-0.2634 0.9256 -0.2420 0.1241
0.5813 0.0211 -0.1409 0.8012
0.5656 0.0654 -0.6338 -0.5235
readtable() | table2array() | pca()
5.2 Split data into training and testing data and export as a .csv file.
sizeIris = size(iris); numRows = sizeIris(1); % Set the seed of the random number generator % for reproducibility. rng(29); [trainInd, valInd, testInd] = dividerand(numRows, 0.7, 0, 0.3); train = iris(trainInd,:); test = iris(testInd,:); csvwrite('iris_train_ML.csv', table2array(train)); csvwrite('iris_test_ML.csv', table2array(test));size() | rng() | dividerand() | table2array() | csvwrite()
5.3 Fit a logistic regression model.
formatSpec = '%d%f%f%C%C%C%C%d'; tips = readtable('tips.csv', 'Delimiter', ',', ... 'Format', formatSpec); tips.fifteen = 0.15 * tips.total_bill; tips.greater15 = (tips.tip > tips.fifteen); [b, dev, stats] = glmfit(tips.total_bill, tips.greater15, 'binomial', 'link', 'logit'); fprintf('The coefficients of the model are: %.3f and %.3f\n', b(1), b(2)); fprintf('The deviance of the fit of the fit is: %.3f\n', dev); fprintf('Other statistics of the model are:\n'); disp(stats);
The coefficients of the model are: 1.648 and -0.072
The deviance of the fit of the fit is: 313.743
Other statistics of the model are:
beta: [2×1 double]
dfe: 242
sfit: 1.0097
s: 1
estdisp: 0
covb: [2×2 double]
se: [2×1 double]
coeffcorr: [2×2 double]
t: [2×1 double]
p: [2×1 double]
resid: [244×1 double]
residp: [244×1 double]
residd: [244×1 double]
resida: [244×1 double]
wts: [244×1 double]
readtable() | glmfit() | fprintf()
5.4 Fit a linear regression model.
linreg = fitlm(tips,'tip~total_bill');
disp(linreg);
Linear regression model:
tip ~ 1 + total_bill
Estimated Coefficients:
Estimate SE tStat pValue
________ _________ ______ __________
(Intercept) 0.92027 0.15973 5.7612 2.5264e-08
total_bill 0.10502 0.0073648 14.26 6.6925e-34
Number of observations: 244, Error degrees of freedom: 242
Root Mean Squared Error: 1.02
R-squared: 0.457, Adjusted R-Squared 0.454
F-statistic vs. constant model: 203, p-value = 6.69e-34
fitlm()
6 Supervised Machine Learning
6.1 Fit a logistic regression model on training data and assess against testing data.
a) Fit a logistic regression model on training data.
formatSpec = '%f%d%d%d%f'; train = readtable('tips_train.csv', 'Delimiter', ',', ... 'Format', formatSpec); test = readtable('tips_test.csv', 'Delimiter', ',', ... 'Format', formatSpec); train.fifteen = 0.15 * train.total_bill; train.greater15 = (train.tip > train.fifteen); test.fifteen = 0.15 * test.total_bill; test.greater15 = (test.tip > test.fifteen); [b, dev, stats] = glmfit(train.total_bill, train.greater15, 'binomial', 'link', 'logit'); fprintf('The coefficients of the model are: %.3f and %.3f\n', b(1), b(2)); fprintf('The deviance of the fit of the fit is: %.3f\n', dev); fprintf('Other statistics of the model are:\n'); disp(stats);
The coefficients of the model are: 1.646 and -0.071
The deviance of the fit of the fit is: 250.584
Other statistics of the model are:
beta: [2×1 double]
dfe: 193
sfit: 1.0107
s: 1
estdisp: 0
covb: [2×2 double]
se: [2×1 double]
coeffcorr: [2×2 double]
t: [2×1 double]
p: [2×1 double]
resid: [195×1 double]
residp: [195×1 double]
residd: [195×1 double]
resida: [195×1 double]
wts: [195×1 double]
readtable() | glmfit() | fprintf()
b) Assess the model against the testing data.
predictions = glmval(b, test.total_bill, 'logit'); predY = round(predictions); Results = strings(size(test,1),1); for i = 1:size(test,1) if (predY(i) == test.greater15(i)) Results(i) = 'Correct'; else Results(i) = 'Wrong'; end end tabulate(Results);
Value Count Percent
Correct 34 69.39%
Wrong 15 30.61%
glmval() | round() | size() | strings() | tabulate()
6.2 Fit a linear regression model on training data and assess against testing data.
a) Fit a linear regression model on training data.
train = readtable('boston_train.xlsx'); test = readtable('boston_test.xlsx'); x_train = train; x_train.Target = []; y_train = train.Target; x_test = test; x_test.Target = []; y_test = test.Target; linreg = fitlm(table2array(x_train), y_train); disp(linreg);
Linear regression model:
y ~ 1 + x1 + x2 + x3 + x4 + x5 + x6 + x7 + x8 + x9 + x10 + x11 + x12 + x13
Estimated Coefficients:
Estimate SE tStat pValue
__________ _________ ________ __________
(Intercept) 36.108 6.505 5.5509 5.7321e-08
x1 -0.085634 0.042774 -2.002 0.046077
x2 0.046034 0.01715 2.6842 0.0076262
x3 0.036413 0.076006 0.47909 0.63219
x4 3.248 1.0741 3.0238 0.0026862
x5 -14.873 4.6361 -3.2081 0.0014633
x6 3.5769 0.53699 6.6609 1.0962e-10
x7 -0.0087032 0.016853 -0.51643 0.60589
x8 -1.3689 0.25296 -5.4115 1.1818e-07
x9 0.31312 0.082366 3.8016 0.00017037
x10 -0.012882 0.0045986 -2.8012 0.0053829
x11 -0.9769 0.171 -5.713 2.4255e-08
x12 0.011326 0.0033585 3.3722 0.00083155
x13 -0.52672 0.062563 -8.419 1.0751e-15
Number of observations: 354, Error degrees of freedom: 340
Root Mean Squared Error: 4.99
R-squared: 0.724, Adjusted R-Squared 0.713
F-statistic vs. constant model: 68.5, p-value = 1.39e-86
readtable() | fitlm()
b) Assess the model against the testing data.
predictions = predict(linreg, table2array(x_test)); sq_diff = (predictions - y_test) .^ 2; disp(mean(sq_diff));
17.7713
6.3 Fit a decision tree model on training data and assess against testing data.
a) Fit a decision tree classification model.
i) Fit a decision tree classification model on training data and determine variable importance.
train = readtable('breastcancer_train.xlsx'); test = readtable('breastcancer_test.xlsx'); x_train = train; x_train.Target = []; y_train = train.Target; x_test = test; x_test.Target = []; y_test = test.Target; rng(29); treeMod = fitctree(x_train, y_train); var_import = predictorImportance(treeMod); var_import = var_import'; var_import(:,2) = var_import; for i = 1:size(var_import,1) var_import(i,1) = i; end var_import = sortrows(var_import, 2, 'descend'); disp(var_import(1:5,:));
24.0000 0.0292
28.0000 0.0077
2.0000 0.0019
12.0000 0.0013
5.0000 0.0009
readtable() | rng() | fitctree() | predictorImportance() | size() | sortrows()
ii) Assess the model against the testing data.
predictions = predict(treeMod, x_test); Results = strings(size(test,1),1); for i = 1:size(test,1) if (predictions(i) == y_test(i)) Results(i) = 'Correct'; else Results(i) = 'Wrong'; end end tabulate(Results);
Value Count Percent
Correct 160 93.57%
Wrong 11 6.43%
predict() | strings() | size() | tabulate()
b) Fit a decision tree regression model.
i) Fit a decision tree regression model on training data and determine variable importance.
train = readtable('boston_train.xlsx'); test = readtable('boston_test.xlsx'); x_train = train; x_train.Target = []; y_train = train.Target; x_test = test; x_test.Target = []; y_test = test.Target; rng(29); treeMod = fitrtree(x_train, y_train); var_import = predictorImportance(treeMod); var_import = var_import'; var_import(:,2) = var_import; for i = 1:size(var_import,1) var_import(i,1) = i; end var_import = sortrows(var_import, 2, 'descend'); disp(var_import(1:5,:));
6.0000 0.6921
13.0000 0.2451
8.0000 0.1227
5.0000 0.0506
1.0000 0.0359
readtable() | rng() | fitrtree() | predictorImportance() | size() | sortrows()
6.4 Fit a random forest model on training data and assess against testing data.
a) Fit a random forest classification model.
i) Fit a random forest classification model on training data and determine variable importance.
train = readtable('breastcancer_train.xlsx'); test = readtable('breastcancer_test.xlsx'); x_train = train; x_train.Target = []; y_train = train.Target; x_test = test; x_test.Target = []; y_test = test.Target; rng(29); rfMod = fitrensemble(table2array(x_train), y_train, 'Method', 'bag'); var_import = predictorImportance(rfMod); var_import = var_import'; var_import(:,2) = var_import; for i = 1:size(var_import,1) var_import(i,1) = i; end var_import = sortrows(var_import, 2, 'descend'); disp(var_import(1:5,:));
23.0000 0.0045
28.0000 0.0041
24.0000 0.0038
8.0000 0.0029
21.0000 0.0026
readtable() | rng() | table2array() | fitrensemble() | predictorImportance() | size() | sortrows()
ii) Assess the model against the testing data.
predictions = predict(rfMod, table2array(x_test)); predictions = round(predictions); Results = strings(size(test,1),1); for i = 1:size(test,1) if (predictions(i) == y_test(i)) Results(i) = 'Correct'; else Results(i) = 'Wrong'; end end tabulate(Results);
Value Count Percent
Correct 166 97.08%
Wrong 5 2.92%
table2array() | predict() | round() | size() | strings() | tabulate()
b) Fit a random forest regression model.
i) Fit a random forest regression model on training data and determine variable importance.
train = readtable('boston_train.xlsx'); test = readtable('boston_test.xlsx'); x_train = train; x_train.Target = []; y_train = train.Target; x_test = test; x_test.Target = []; y_test = test.Target; rng(29); rfMod = fitrensemble(table2array(x_train), y_train, 'Method', 'bag'); var_import = predictorImportance(rfMod); var_import = var_import'; var_import(:,2) = var_import; for i = 1:size(var_import,1) var_import(i,1) = i; end var_import = sortrows(var_import, 2, 'descend'); disp(var_import(1:5,:));
6.0000 0.4811
13.0000 0.4738
1.0000 0.0873
3.0000 0.0842
8.0000 0.0723
readtable() | rng() | table2array() | fitrensemble() | predictorImportance() | size() | sortrows()
ii) Assess the model against the testing data.
predictions = predict(rfMod, table2array(x_test)); sq_diff = (predictions - y_test) .^ 2; disp(mean(sq_diff));
10.5583table2array() | predict() | mean()
6.7 Fit a support vector model on training data and assess against testing data.
a) Fit a support vector classification model.
i) Fit a support vector classification model on training data.
Note: In implementation scaling should be used.
train = readtable('breastcancer_train.xlsx'); test = readtable('breastcancer_test.xlsx'); x_train = train; x_train.Target = []; y_train = train.Target; x_test = test; x_test.Target = []; y_test = test.Target; rng(29); svMod = fitcsvm(x_train, y_train);readtable() | rng() | fitcsvm()
ii) Assess the model against the testing data.
predictions = predict(svMod, x_test); Results = strings(size(test,1),1); for i = 1:size(test,1) if (predictions(i) == y_test(i)) Results(i) = 'Correct'; else Results(i) = 'Wrong'; end end tabulate(Results);
Value Count Percent
Correct 163 95.32%
Wrong 8 4.68%
predict() | size() | strings() | tabulate()
b) Fit a support vector regression model.
i) Fit a support vector regression model on training data.
Note: In implementation scaling should be used.
train = readtable('boston_train.xlsx'); test = readtable('boston_test.xlsx'); x_train = train; x_train.Target = []; y_train = train.Target; x_test = test; x_test.Target = []; y_test = test.Target; rng(29); svMod = fitrsvm(x_train, y_train);readtable() | rng() | fitrsvm()
7 Unsupervised Machine Learning
7.1 KMeans Clustering
formatSpec = '%f%f%f%f%d'; iris = readtable('iris.csv', 'Delimiter', ',', ... 'Format', formatSpec); features = table2array(iris(:, {'SepalLength', 'SepalWidth', 'PetalLength', 'PetalWidth'})); iris.Labels = strings(size(iris,1),1); for i = 1:size(iris,1) if (iris.Target(i) == 0) iris.Labels(i) = 'Setosa'; else if (iris.Target(i) == 1) iris.Labels(i) = 'Versicolor'; else iris.Labels(i) = 'Virginica'; end end end rng(29); [labels, C] = kmeans(features, 3); iris.Predictions = labels; disp(crosstab(iris.Labels, iris.Predictions));
0 50 0
3 0 47
36 0 14
readtable() | table2array() | size() | strings() | kmeans() | crosstab()
7.3 Ward Hierarchical Clustering
rng(29); tree = linkage(features, 'ward', 'euclidean', 'savememory', 'on'); labels = cluster(tree, 'maxclust', 3); iris.Predictions = labels; disp(crosstab(iris.Labels, iris.Predictions));
0 0 50
1 49 0
35 15 0
rng() | linkage() | cluster() | crosstab()
8 Forecasting
8.1 Fit an ARIMA model to a timeseries.
a) Plot the timeseries.
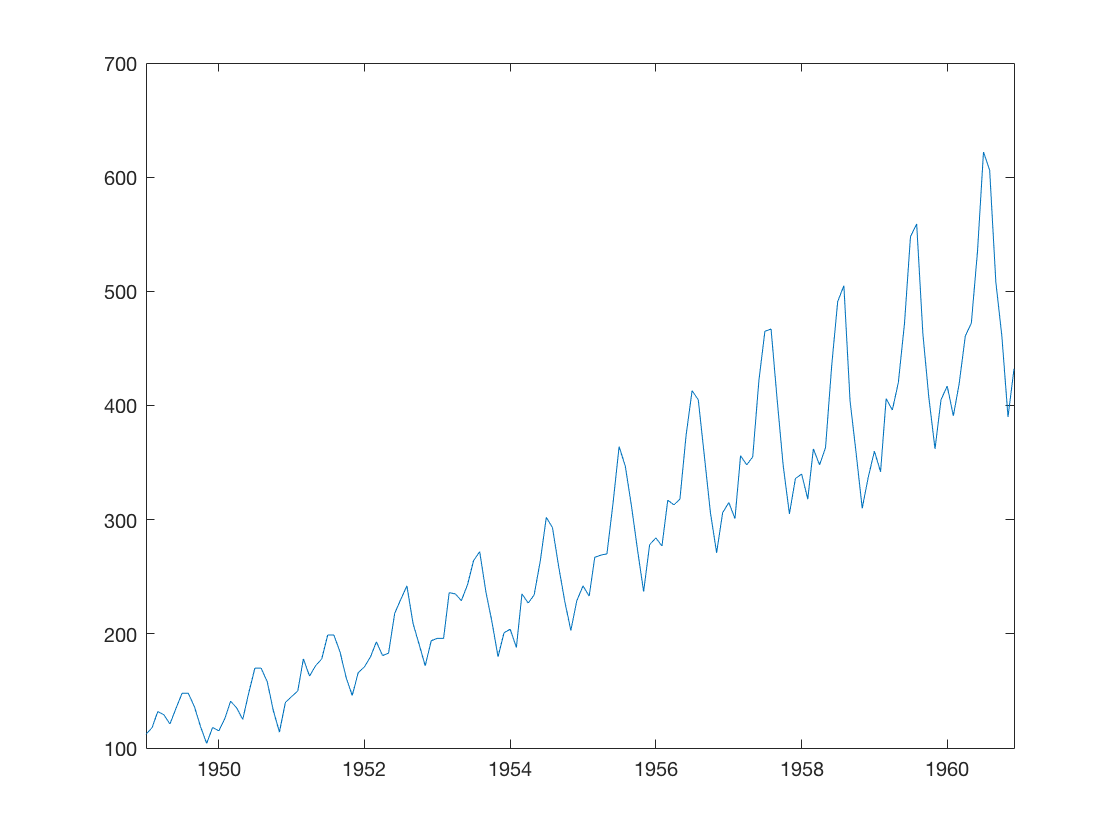
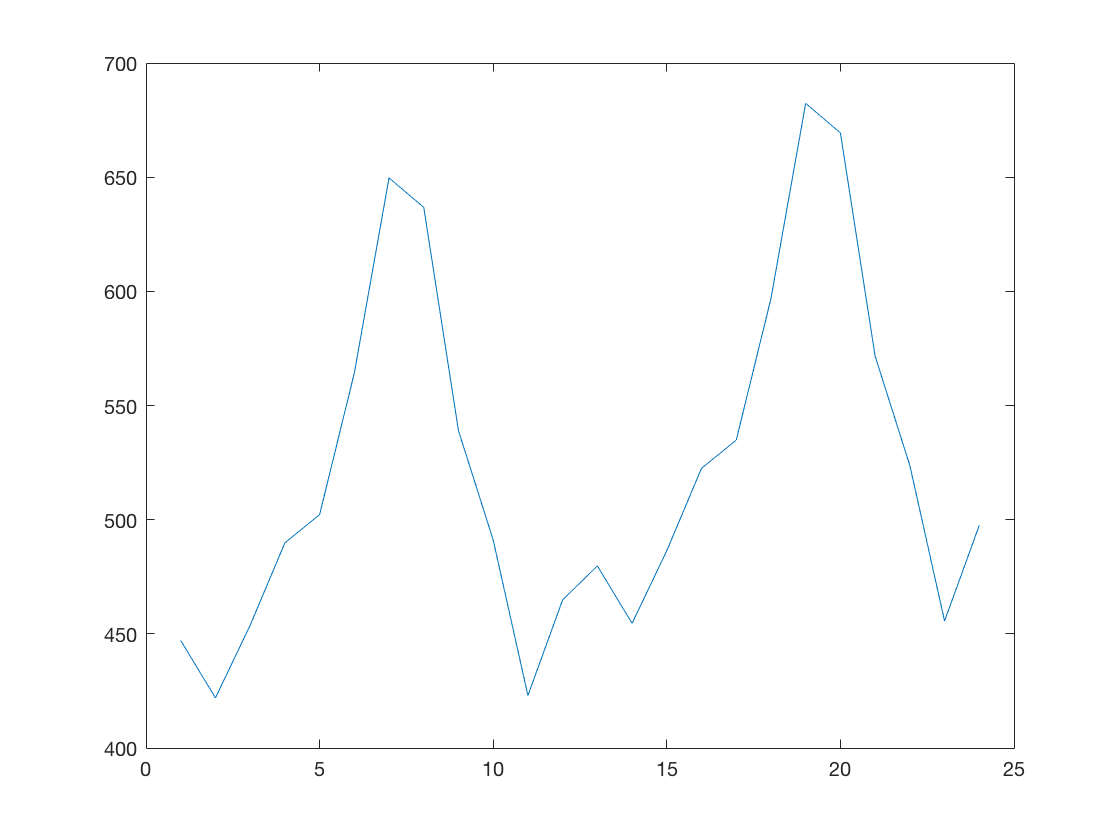
b) Fit an ARIMA model and predict 2 years (24 months).
model = arima('Constant',0,'D',1,'Seasonality',12,... 'MALags',1,'SMALags',12); est_model = estimate(model, air.AIR); [yF, yMSE] = forecast(est_model, 24, 'Y0', air.AIR); plot(yF);
ARIMA(0,1,1) Model Seasonally Integrated with Seasonal MA(12):
---------------------------------------------------------------
Conditional Probability Distribution: Gaussian
Standard t
Parameter Value Error Statistic
----------- ----------- ------------ -----------
Constant 0 Fixed Fixed
MA{1} -0.309349 0.0619375 -4.99454
SMA{12} -0.112821 0.082419 -1.36887
Variance 123.282 12.5426 9.82908
9 Model Evaluation & Selection
9.1 Evaluate the accuracy of regression models.
a) Evaluation on training data.
train = readtable('boston_train.xlsx'); test = readtable('boston_test.xlsx'); x_train = train; x_train.Target = []; y_train = train.Target; x_test = test; x_test.Target = []; y_test = test.Target; rng(29); rfMod = fitrensemble(table2array(x_train), y_train, 'Method', 'bag'); predictions = predict(rfMod, table2array(x_train)); r2_rf = 1 - ( (sum((y_train - predictions) .^ 2)) / (sum((y_train - mean(y_train)) .^ 2)) ); fprintf('Random forest regression model r^2 score (coefficient of determination): %.3f\n', r2_rf);
Random forest regression model r^2 score (coefficient of determination): 0.930readtable() | rng() | table2array() | fitrensemble() | predict() | mean() | sum() | fprintf()
b) Evaluation on testing data.
predictions = predict(rfMod, table2array(x_test));
r2_rf = 1 - ( (sum((y_test - predictions) .^ 2)) / (sum((y_test - mean(y_test)) .^ 2)) );
fprintf('Random forest regression model r^2 score (coefficient of determination): %.3f\n', r2_rf);
Random forest regression model r^2 score (coefficient of determination): 0.867predict() | mean() | sum() | fprintf()
9.2 Evaluate the accuracy of classification models.
a) Evaluation on training data.
train = readtable('digits_train.xlsx'); test = readtable('digits_test.xlsx'); x_train = train; x_train.Target = []; y_train = train.Target; x_test = test; x_test.Target = []; y_test = test.Target; rng(29); rfMod = fitrensemble(table2array(x_train), y_train, 'Method', 'bag'); predY = predict(rfMod, table2array(x_train)); predY = round(predY); Results = zeros(size(train,1),1); for i = 1:size(Results,1) if (predY(i) == y_train(i)) Results(i) = 1; else Results(i) = 0; end end accuracy_rf = (1/size(x_train,1)) * sum(Results); fprintf('Random forest model accuracy: %.3f\n', accuracy_rf);
Random forest model accuracy: 0.692readtable() | rng() | table2array() | fitrensemble() | predict() | round() | size() | zeros() | sum() | fprintf()
b) Evaluation on testing data.
predY = predict(rfMod, table2array(x_test)); predY = round(predY); Results = zeros(size(test,1),1); for i = 1:size(Results,1) if (predY(i) == y_test(i)) Results(i) = 1; else Results(i) = 0; end end accuracy_rf = (1/size(x_test,1)) * sum(Results); fprintf('Random forest model accuracy: %.3f\n', accuracy_rf);
Random forest model accuracy: 0.526predict() | round() | size() | zeros() | sum() | fprintf()
Appendix
1 Built-in MATLAB Data Types
Numeric types
- Integer and floating-point data
Characters and strings
- Text in character arrays and string arrays
Tables
- Arrays in tabular form whose named columns can have different types
Structures
- Arrays with named fields that can contain data of varying types and sizes
Cell arrays
- Arrays that can contain data of varying types and sizes
Alphabetical Index
Note, from the MathWorks website: "MATLAB is an abbreviation for "matrix laboratory." While other programming languages mostly work with numbers one at a time, MATLAB® is designed to operate primarily on whole matrices and arrays. All MATLAB variables are multidimensional arrays, no matter what type of data."
Matrix
A matrix is a two-dimensional array often used for linear algebra operations. Please see the following example of matrix creation and access:
my_matrix = [1 2 3; 4 5 6; 7 8 9] disp(my_matrix(2,2));
5